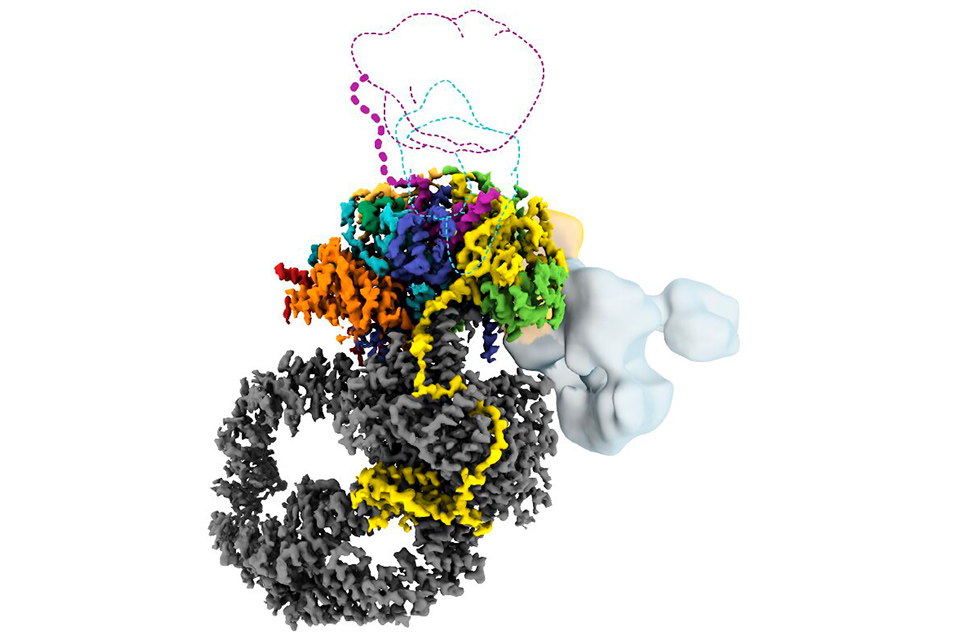
CHEVY CHASE, MD.- For the first time, researchers have constructed a detailed 3D structure of human SAGA, a 20-piece molecular machine crucial to human life.
Although errors in this protein complex contribute to distinctly human problems, including autism, neurological disorders, and cancers such as melanoma, SAGA research has long focused on the yeast version. It's simpler than human SAGA and easier to study; last year scientists reported yeast SAGA's structure.
"We often look at the molecular machinery of organisms like yeast to understand how it functions in humans and how errors can cause disease," says
Howard Hughes Medical Institute Investigator Eva Nogales, a structural biologist at the University of California, Berkeley. But she and her team suspected they would find key differences between the yeast and human structures.
For one thing, Nogales says, problems with SAGA affect mammals and yeast quite differently. Without SAGA, for example, mouse embryos fail to develop, while yeast cells find a way to compensate. What's more, researchers had already recognized differences in the proteins contained in the yeast versus human complex.
And in fact, Nogales's team reports in the journal Nature Structural & Molecular Biology on November 22, 2021, the human SAGA structure revealed dramatic changes from its yeast counterpart. Critical pieces of the complex had been completely rearranged, Nogales, Berkeley HHMI Investigator Robert Tjian, and their colleagues discovered.
"We know a lot about the yeast SAGA complex, but in humans, it's kind of a murky business," says Steven Hahn, a molecular biologist at the Fred Hutchinson Cancer Research Center who was not involved in the study. "This new structure is really going to inform a lot of experiments going forward."
From controlling traffic into the nucleus to copying DNA, some tasks within cells require a team effort. To get these and other jobs done, multiple proteins assemble into physical structures. Among such protein complexes, human SAGA is about as big as they come, with 20 component proteins. It belongs to a group of large complexes that are "big and floppy, with multiple lobes that all move with respect to each other," says Hahn, who studies SAGA and other molecular machines like it.
SAGA uses its appendages to help control access to genes packed within chromosomes. It also plays a key role in activating transcription, the first step of the process by which genetic information is ultimately converted into proteins. Genetic mutations that interfere with SAGA's ability to perform these tasks can cause disease.
Examining the architecture of this complex could help scientists understand how such mistakes disrupt SAGA's duties, potentially opening the door to treatments. But mapping SAGA's structure—in yeast or humans—wasn't easy. Not only is the complex large, but its flexible parts make it difficult to visualize using conventional methods. Instead, Nogales's team relied on a technology called cryo-electron microscopy, in which a sample is rapidly cooled and then blasted with electrons to image molecules in action.
Human SAGA is difficult to obtain in large enough quantities for cryo-EM. So, Nogales's team genetically engineered a version of SAGA with molecular handles. The researchers used these handles to pull SAGA from lab-grown cells, then examined the complex with cryo-EM.
Raw images of the structure started out fuzzy, she says, like a view on a foggy day. Computational processing improved the resolution in stages. "It's like lifting the fog," says Nogales, who has been examining molecular structures for nearly 30 years. "It was very exciting to see what was actually there, especially the elements we didn't expect."
The final structure the team generated revealed how evolution had tinkered with human SAGA, positioning two major modules in a different orientation compared to yeast, for example. The team also discovered a small component threaded through the human complex that acts as a latch securing one of SAGA's mobile parts.
Nogales and her team used their new structure to map the locations of the 13 known SAGA mutations linked to disease. With this physical guide, scientists can better examine the effects of these errors and, potentially, develop drugs to restore SAGA's function.
While in most cases the researchers don't yet know how the mutations hinder SAGA, two involve the latch component that Nogales's team discovered. "If we had tried to map these mutations onto a yeast structure, it wouldn't have told us anything about what is going on in disease," Nogales says.